Note
Go to the end to download the full example code
Compute Eigenvalues using MAPDL or SciPy#
This example shows a comparison between MAPDL and Scipy eigenvalues computation power.
More specificatly, this example shows:
How to extract the stiffness and mass matrices from a MAPDL model.
How to use the
Mathmodule of PyMapdl to compute the first eigenvalues.How to can get these matrices in the SciPy world, to get the same solutions using Python resources.
How MAPDL is really faster than SciPy :)
import math
Model setup#
First load python packages we need for this example
import time
import matplotlib.pylab as plt
import numpy as np
import scipy
from scipy.sparse.linalg import eigsh
from ansys.mapdl.core import examples, launch_mapdl
Next:
Load the ansys.mapdl module
Get the
Mathmodule of PyMapdl
mapdl = launch_mapdl()
print(mapdl)
mm = mapdl.math
Product: Ansys Mechanical Enterprise
MAPDL Version: 23.1
ansys.mapdl Version: 0.65.2
APDLMath EigenSolve First load the input file using MAPDL.
print(mapdl.input(examples.examples.wing_model))
/INPUT FILE= LINE= 0
*****MAPDL VERIFICATION RUN ONLY*****
DO NOT USE RESULTS FOR PRODUCTION
***** MAPDL ANALYSIS DEFINITION (PREP7) *****
*** WARNING *** CP = 0.000 TIME= 00:00:00
Deactivation of element shape checking is not recommended.
*** WARNING *** CP = 0.000 TIME= 00:00:00
The mesh of area 1 contains PLANE42 triangles, which are much too stiff
in bending. Use quadratic (6- or 8-node) elements if possible.
*** WARNING *** CP = 0.000 TIME= 00:00:00
CLEAR, SELECT, and MESH boundary condition commands are not possible
after MODMSH.
***** ROUTINE COMPLETED ***** CP = 0.000
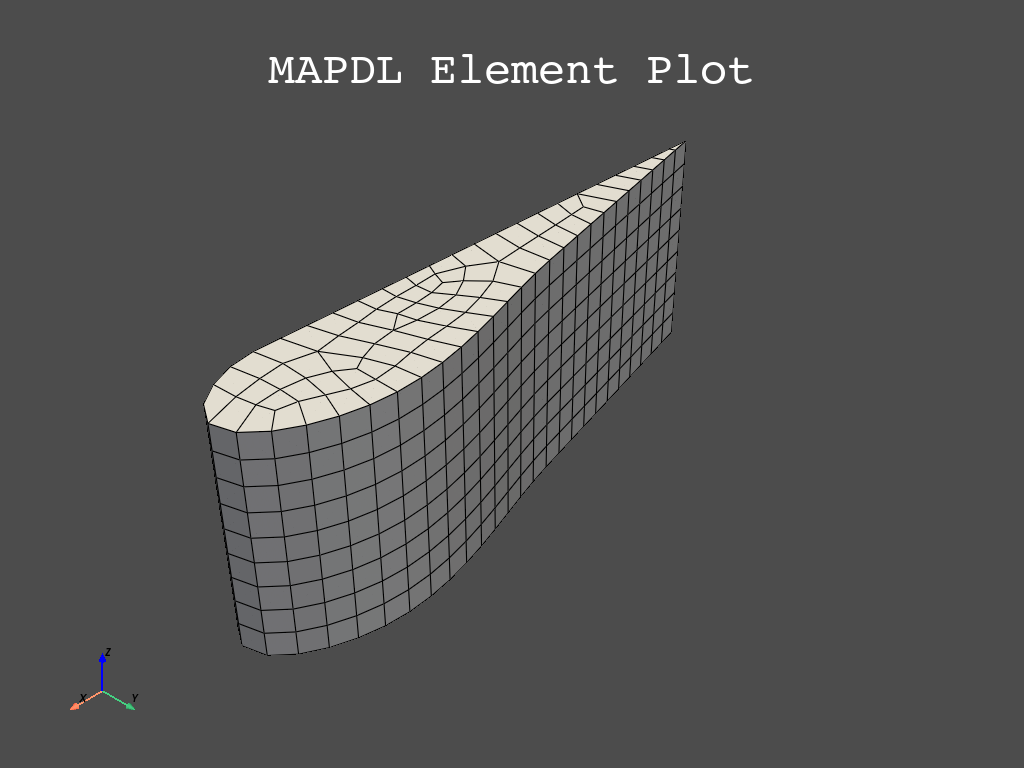
Plot and mesh using the eplot method.
mapdl.eplot()
Next, setup a modal Analysis and request the .FULL
file.
print(mapdl.slashsolu())
print(mapdl.antype(antype="MODAL"))
print(mapdl.modopt(method="LANB", nmode="10", freqb="1."))
print(mapdl.wrfull(ldstep="1"))
# store the output of the solve command
output = mapdl.solve()
***** MAPDL SOLUTION ROUTINE *****
PERFORM A MODAL ANALYSIS
THIS WILL BE A NEW ANALYSIS
USE SYM. BLOCK LANCZOS MODE EXTRACTION METHOD
EXTRACT 10 MODES
SHIFT POINT FOR EIGENVALUE CALCULATION= 1.0000
NORMALIZE THE MODE SHAPES TO THE MASS MATRIX
STOP SOLUTION AFTER FULL FILE HAS BEEN WRITTEN
LOADSTEP = 1 SUBSTEP = 1 EQ. ITER = 1
Matrices manipulation using PyMAPDL#
Read the sparse matrices using PyMapdl.
mapdl.finish()
mm.free()
k = mm.stiff(fname="file.full")
M = mm.mass(fname="file.full")
Solve the eigenproblem using PyMapdl with APDLMath.
Elapsed time to solve this problem : 0.6976687908172607
Print eigenfrequencies and accuracy.
Accuracy :
mapdl_acc = np.empty(nev)
for i in range(nev):
f = ev[i] # Eigenfrequency (Hz)
omega = 2 * np.pi * f # omega = 2.pi.Frequency
lam = omega**2 # lambda = omega^2
phi = A[i] # i-th eigenshape
kphi = k.dot(phi) # K.Phi
mphi = M.dot(phi) # M.Phi
kphi_nrm = kphi.norm() # Normalization scalar value
mphi *= lam # (K-\lambda.M).Phi
kphi -= mphi
mapdl_acc[i] = kphi.norm() / kphi_nrm # compute the residual
print(f"[{i}] : Freq = {f:8.2f} Hz\t Residual = {mapdl_acc[i]:.5}")
[0] : Freq = 352.39 Hz Residual = 1.3931e-08
[1] : Freq = 385.16 Hz Residual = 2.3293e-08
[2] : Freq = 656.78 Hz Residual = 3.9369e-09
[3] : Freq = 764.72 Hz Residual = 9.5144e-09
[4] : Freq = 825.33 Hz Residual = 7.3415e-09
[5] : Freq = 1039.21 Hz Residual = 4.9065e-09
[6] : Freq = 1143.66 Hz Residual = 1.7584e-08
[7] : Freq = 1258.00 Hz Residual = 5.1418e-08
[8] : Freq = 1334.23 Hz Residual = 5.7417e-08
[9] : Freq = 1352.06 Hz Residual = 4.1036e-08
Use SciPy to Solve the same Eigenproblem#
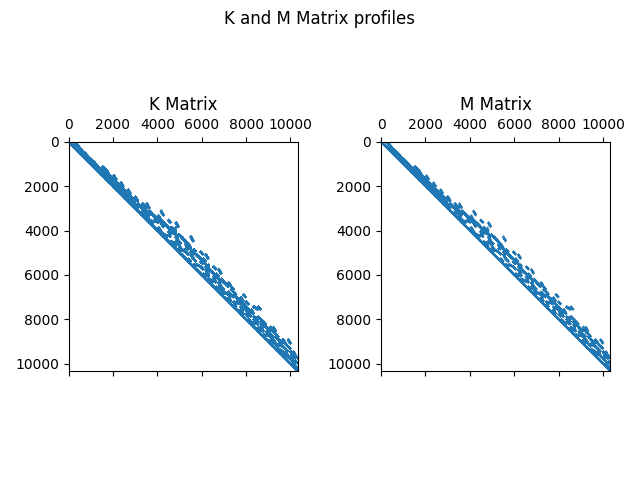
First get MAPDL sparse matrices into the Python memory as SciPy matrices.
pk = k.asarray()
pm = M.asarray()
# get_ipython().run_line_magic('matplotlib', 'inline')
fig, (ax1, ax2) = plt.subplots(1, 2)
fig.suptitle("K and M Matrix profiles")
ax1.spy(pk, markersize=0.01)
ax1.set_title("K Matrix")
ax2.spy(pm, markersize=0.01)
ax2.set_title("M Matrix")
plt.tight_layout()
plt.show(block=True)
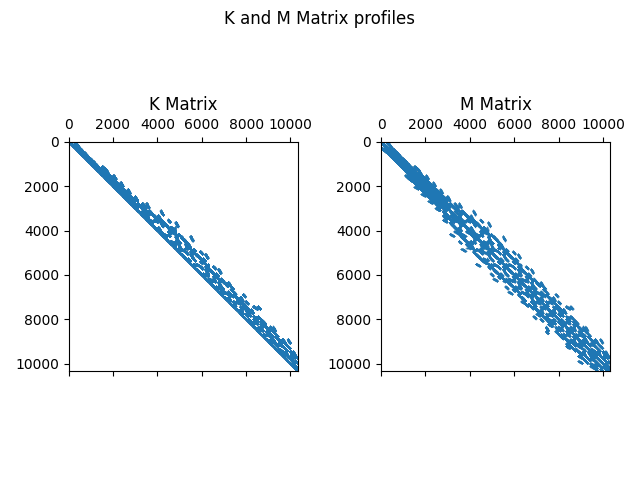
Make the sparse matrices for SciPy unsymmetric as symmetric matrices in SciPy are memory inefficient.
pkd = scipy.sparse.diags(pk.diagonal())
pK = pk + pk.transpose() - pkd
pmd = scipy.sparse.diags(pm.diagonal())
pm = pm + pm.transpose() - pmd
Plot Matrices
fig, (ax1, ax2) = plt.subplots(1, 2)
fig.suptitle("K and M Matrix profiles")
ax1.spy(pk, markersize=0.01)
ax1.set_title("K Matrix")
ax2.spy(pm, markersize=0.01)
ax2.set_title("M Matrix")
plt.tight_layout()
plt.show(block=True)
Solve the eigenproblem
Elapsed time to solve this problem : 3.766496419906616
Convert Lambda values to Frequency values:
Compute the residual error for SciPy.
scipy_acc = np.zeros(nev)
for i in range(nev):
lam = vals[i] # i-th eigenvalue
phi = vecs.T[i] # i-th eigenshape
kphi = pk * phi.T # K.Phi
mphi = pm * phi.T # M.Phi
kphi_nrm = np.linalg.norm(kphi, 2) # Normalization scalar value
mphi *= lam # (K-\lambda.M).Phi
kphi -= mphi
scipy_acc[i] = 1 - np.linalg.norm(kphi, 2) / kphi_nrm # compute the residual
print(f"[{i}] : Freq = {freqs[i]:8.2f} Hz\t Residual = {scipy_acc[i]:.5}")
[0] : Freq = 352.39 Hz Residual = 8.0098e-05
[1] : Freq = 385.16 Hz Residual = 0.00010369
[2] : Freq = 656.78 Hz Residual = 0.00024262
[3] : Freq = 764.72 Hz Residual = 0.00016258
[4] : Freq = 825.33 Hz Residual = 0.00039002
[5] : Freq = 1039.21 Hz Residual = 0.00057628
[6] : Freq = 1143.66 Hz Residual = 0.0025916
[7] : Freq = 1258.00 Hz Residual = 0.00033872
[8] : Freq = 1334.22 Hz Residual = 0.00046633
[9] : Freq = 1352.06 Hz Residual = 0.0011262
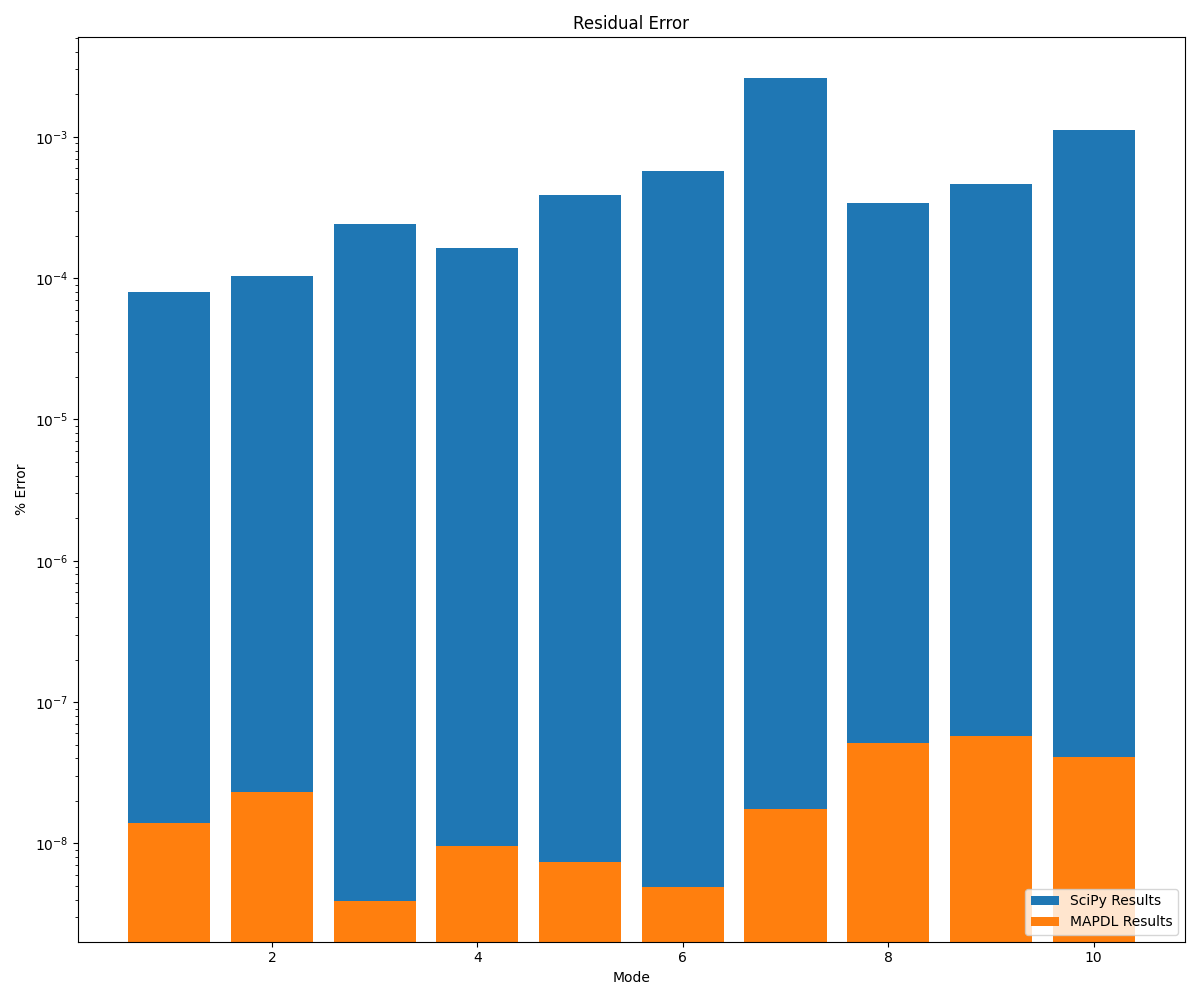
MAPDL vs Scipy comparison#
MAPDL is more accurate than SciPy.
fig = plt.figure(figsize=(12, 10))
ax = plt.axes()
x = np.linspace(1, 10, 10)
plt.title("Residual Error")
plt.yscale("log")
plt.xlabel("Mode")
plt.ylabel("% Error")
ax.bar(x, scipy_acc, label="SciPy Results")
ax.bar(x, mapdl_acc, label="MAPDL Results")
plt.legend(loc="lower right")
plt.tight_layout()
plt.show()
MAPDL is faster than SciPy.
ratio = scipy_elapsed_time / mapdl_elapsed_time
print(f"Mapdl is {ratio:.3} times faster")
Mapdl is 5.4 times faster
Stop mapdl#
mapdl.exit()
Total running time of the script: ( 0 minutes 7.788 seconds)